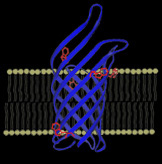
The transmembrane domain of a nonspecific β-barrel pore located in the outer membrane of bacteria, Outer Membrane Protein A (OmpA), is displayed on the left (PDB 1QJP) along with a cartoon representation of the lipid bilayer. The five native tryptophan residues found at the bilayer-water interface are shown in red. OmpA serves as our model β-barrel integral protein for fundamental studies on membrane protein folding.
Elucidating the nature of membrane protein folding would have important impacts in chemistry and biology. Foremost, understanding protein assembly in lipid bilayers would contribute to the growing wealth of knowledge of folding pathways for soluble proteins and similarly encourage theoretical explorations of membrane protein folding. In addition, a more rigorous grasp of noncovalent lipid-protein interactions may be achieved through the study of membrane proteins. The fields of pathology and medicine would greatly benefit as well. Numerous channel-forming bacterial proteins, such as the Staphylococcus aureus &alpha-hemolysis protein and the anthrax toxin protective antigen from Bacillus anthracis transform from a benign water-soluble state to a toxic membrane-bound form. Cell invasion by viruses, such as the influenza virus hemagglutinin, also involves peptide insertion into a membrane. Membrane proteins may undergo aberrant folding or assembly to give rise to numerous common diseases, such as cystic fibrosis and diabetes. Despite the ubiquity and continuously growing library of membrane proteins, there is a dearth of fundamental knowledge of the folding and assembly of these proteins. The aim of our research is to probe molecular mechanisms for membrane protein folding and aggregation with focus on: (1) evolution of secondary structures and aromatic amino acid hydrogen bonding states; (2) changes in local protein solvent environment; and (3) micro- and macroscopic factors that induce protein misfolding and aggregation. The tools utilized for these studies include steady-state and time-resolved resonance emission spectroscopy (fluorescence, phosphorescence, and UV resonance Raman). Return to main research page Read about resonance Raman spectroscopy |